The ezPurrr package is built on top of the purrr library. The purpose of the package is to help make functional programming workflows more simple and efficient. Let’s start by loading ezPurrr and a few other packages we’ll use for data manipulation in this vignette.
ezPurrr requires a nested dataset. For example
nest_df <- palmerpenguins::penguins %>%
group_by(island, species) %>%
nest()
head(nest_df)
#> # A tibble: 5 × 3
#> # Groups: island, species [5]
#> species island data
#> <fct> <fct> <list>
#> 1 Adelie Torgersen <tibble [52 × 6]>
#> 2 Adelie Biscoe <tibble [44 × 6]>
#> 3 Adelie Dream <tibble [56 × 6]>
#> 4 Gentoo Biscoe <tibble [124 × 6]>
#> 5 Chinstrap Dream <tibble [68 × 6]>This data frame has columns for the penguin species and the island on which the data was collected but, critically, an additional data column. This column is a list column that has all the data for the corresponding species/island row. All functions in ezPurrr require the data to first be in this format.
From here, we want to be able to work with the data like normal, but it’s difficult when the data are in this list column format. This is where teh sample_*() functions come in.
sample_*()
sample_row()
The sample_row() function returns one random or one particular row (when an index argument is supplied) from the nested data as a list or a data frame. If returning a list (default), the first item of the list with be the data from the given row, while the remaining elements of the list will be the grouping variables from that row (i.e., the species and island for the corresponding row, in our previous example). If returning a data frame (with type = df argument), then the data frame will include one row of the nested data, as well as each grouping variable as a column (with identical repeated content).
For example
nest_df %>%
sample_row()
#> row 5 is selected randomly
#> $data
#> # A tibble: 68 × 6
#> bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year
#> <dbl> <dbl> <int> <int> <fct> <int>
#> 1 46.5 17.9 192 3500 female 2007
#> 2 50 19.5 196 3900 male 2007
#> 3 51.3 19.2 193 3650 male 2007
#> 4 45.4 18.7 188 3525 female 2007
#> 5 52.7 19.8 197 3725 male 2007
#> 6 45.2 17.8 198 3950 female 2007
#> 7 46.1 18.2 178 3250 female 2007
#> 8 51.3 18.2 197 3750 male 2007
#> 9 46 18.9 195 4150 female 2007
#> 10 51.3 19.9 198 3700 male 2007
#> # … with 58 more rows
#>
#> $island
#> [1] Dream
#> Levels: Biscoe Dream Torgersen
#>
#> $species
#> [1] Chinstrap
#> Levels: Adelie Chinstrap Gentoo
nest_df %>%
sample_row(index = 3) %>%
head()
#> $data
#> # A tibble: 56 × 6
#> bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year
#> <dbl> <dbl> <int> <int> <fct> <int>
#> 1 39.5 16.7 178 3250 female 2007
#> 2 37.2 18.1 178 3900 male 2007
#> 3 39.5 17.8 188 3300 female 2007
#> 4 40.9 18.9 184 3900 male 2007
#> 5 36.4 17 195 3325 female 2007
#> 6 39.2 21.1 196 4150 male 2007
#> 7 38.8 20 190 3950 male 2007
#> 8 42.2 18.5 180 3550 female 2007
#> 9 37.6 19.3 181 3300 female 2007
#> 10 39.8 19.1 184 4650 male 2007
#> # … with 46 more rows
#>
#> $island
#> [1] Dream
#> Levels: Biscoe Dream Torgersen
#>
#> $species
#> [1] Adelie
#> Levels: Adelie Chinstrap Gentoo
nest_df %>%
sample_row(index = 3, type = 'df') %>%
head()
#> # A tibble: 6 × 8
#> bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year
#> <dbl> <dbl> <int> <int> <fct> <int>
#> 1 39.5 16.7 178 3250 female 2007
#> 2 37.2 18.1 178 3900 male 2007
#> 3 39.5 17.8 188 3300 female 2007
#> 4 40.9 18.9 184 3900 male 2007
#> 5 36.4 17 195 3325 female 2007
#> 6 39.2 21.1 196 4150 male 2007
#> # … with 2 more variables: group.island <fct>, group.species <fct>
sample_data()
sample_data() returns one random or one particular data column (with index argument) from the nested data. The primary difference between sample_data() versus sample_row() is that the grouping variables are not returned - just the data. For example:
nest_df %>%
sample_data()
#> row 2 is selected randomly
#> # A tibble: 44 × 6
#> bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year
#> <dbl> <dbl> <int> <int> <fct> <int>
#> 1 37.8 18.3 174 3400 female 2007
#> 2 37.7 18.7 180 3600 male 2007
#> 3 35.9 19.2 189 3800 female 2007
#> 4 38.2 18.1 185 3950 male 2007
#> 5 38.8 17.2 180 3800 male 2007
#> 6 35.3 18.9 187 3800 female 2007
#> 7 40.6 18.6 183 3550 male 2007
#> 8 40.5 17.9 187 3200 female 2007
#> 9 37.9 18.6 172 3150 female 2007
#> 10 40.5 18.9 180 3950 male 2007
#> # … with 34 more rows
nest_df %>%
sample_data(index = 3)
#> # A tibble: 56 × 6
#> bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year
#> <dbl> <dbl> <int> <int> <fct> <int>
#> 1 39.5 16.7 178 3250 female 2007
#> 2 37.2 18.1 178 3900 male 2007
#> 3 39.5 17.8 188 3300 female 2007
#> 4 40.9 18.9 184 3900 male 2007
#> 5 36.4 17 195 3325 female 2007
#> 6 39.2 21.1 196 4150 male 2007
#> 7 38.8 20 190 3950 male 2007
#> 8 42.2 18.5 180 3550 female 2007
#> 9 37.6 19.3 181 3300 female 2007
#> 10 39.8 19.1 184 4650 male 2007
#> # … with 46 more rows
sample_group()
Finally, sample_group() returns one random or one particular grouping columns (with index argument) from the nested dataset with no data. For example:
nest_df %>%
sample_group()
#> row 3 is selected randomly
#> # A tibble: 1 × 2
#> # Groups: island, species [1]
#> species island
#> <fct> <fct>
#> 1 Adelie Dream
nest_df %>%
sample_group(index = 3)
#> # A tibble: 1 × 2
#> # Groups: island, species [1]
#> species island
#> <fct> <fct>
#> 1 Adelie Dream
broadcast()
broadcast()
Generally, we will use the data we have sampled to conduct some operations. Once we’ve conducted those operations we just need to wrap them all in a function, then broadcast() them across the sets of data (i.e., all rows).
In other words, the sampling dataset is used to test the code you want to be applied for each row, such as figures, models, or transformations.
Let’s look at an example for plotting. First, we create our sample data.
samp <- nest_df %>%
sample_data(index = 3)
samp
#> # A tibble: 56 × 6
#> bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year
#> <dbl> <dbl> <int> <int> <fct> <int>
#> 1 39.5 16.7 178 3250 female 2007
#> 2 37.2 18.1 178 3900 male 2007
#> 3 39.5 17.8 188 3300 female 2007
#> 4 40.9 18.9 184 3900 male 2007
#> 5 36.4 17 195 3325 female 2007
#> 6 39.2 21.1 196 4150 male 2007
#> 7 38.8 20 190 3950 male 2007
#> 8 42.2 18.5 180 3550 female 2007
#> 9 37.6 19.3 181 3300 female 2007
#> 10 39.8 19.1 184 4650 male 2007
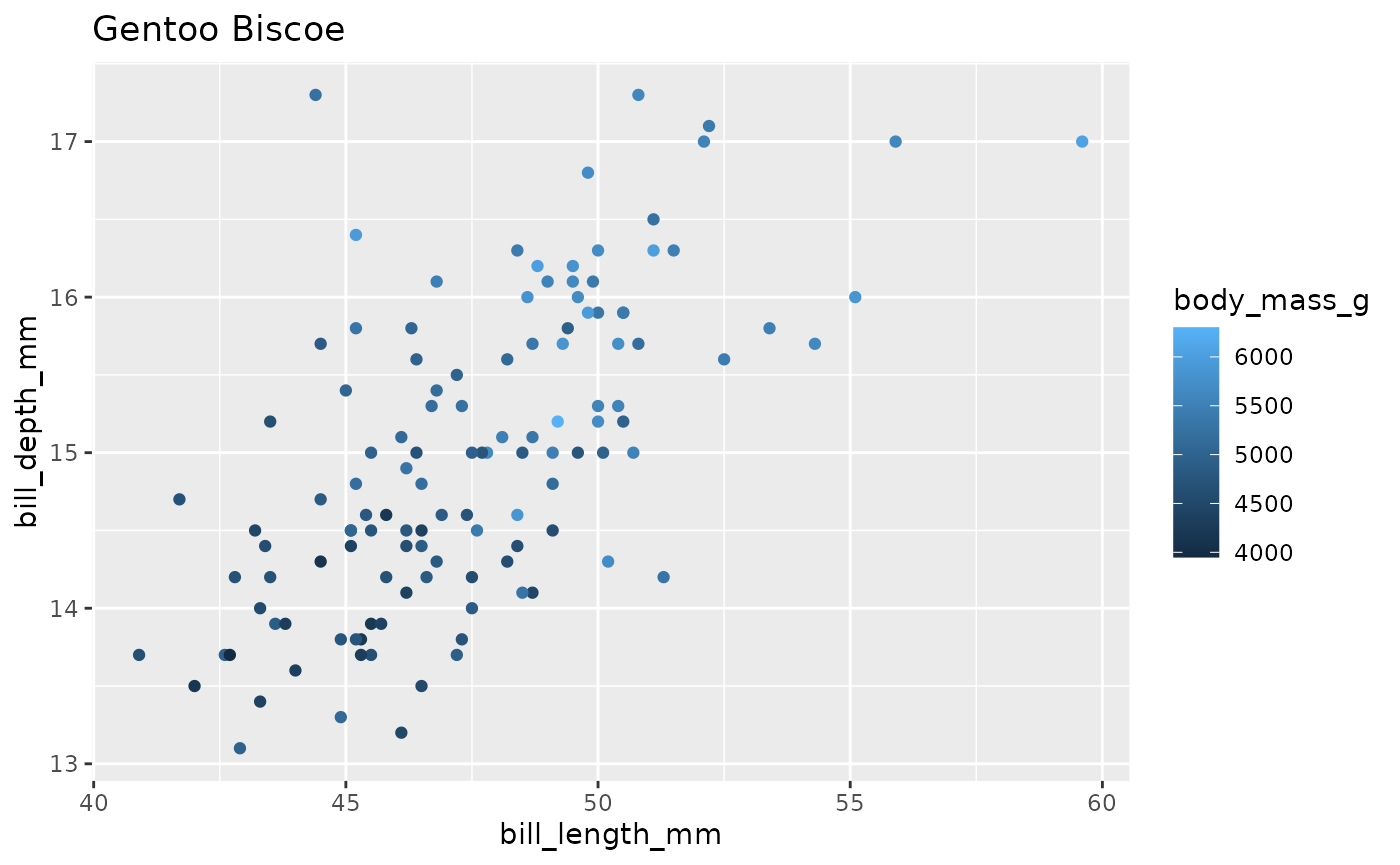
#> # … with 46 more rowsNext, we write the code to create our plot, using samp as our sample data to test our code.
ggplot(samp, aes(x = bill_length_mm,
y = bill_depth_mm,
color = body_mass_g)) +
geom_point()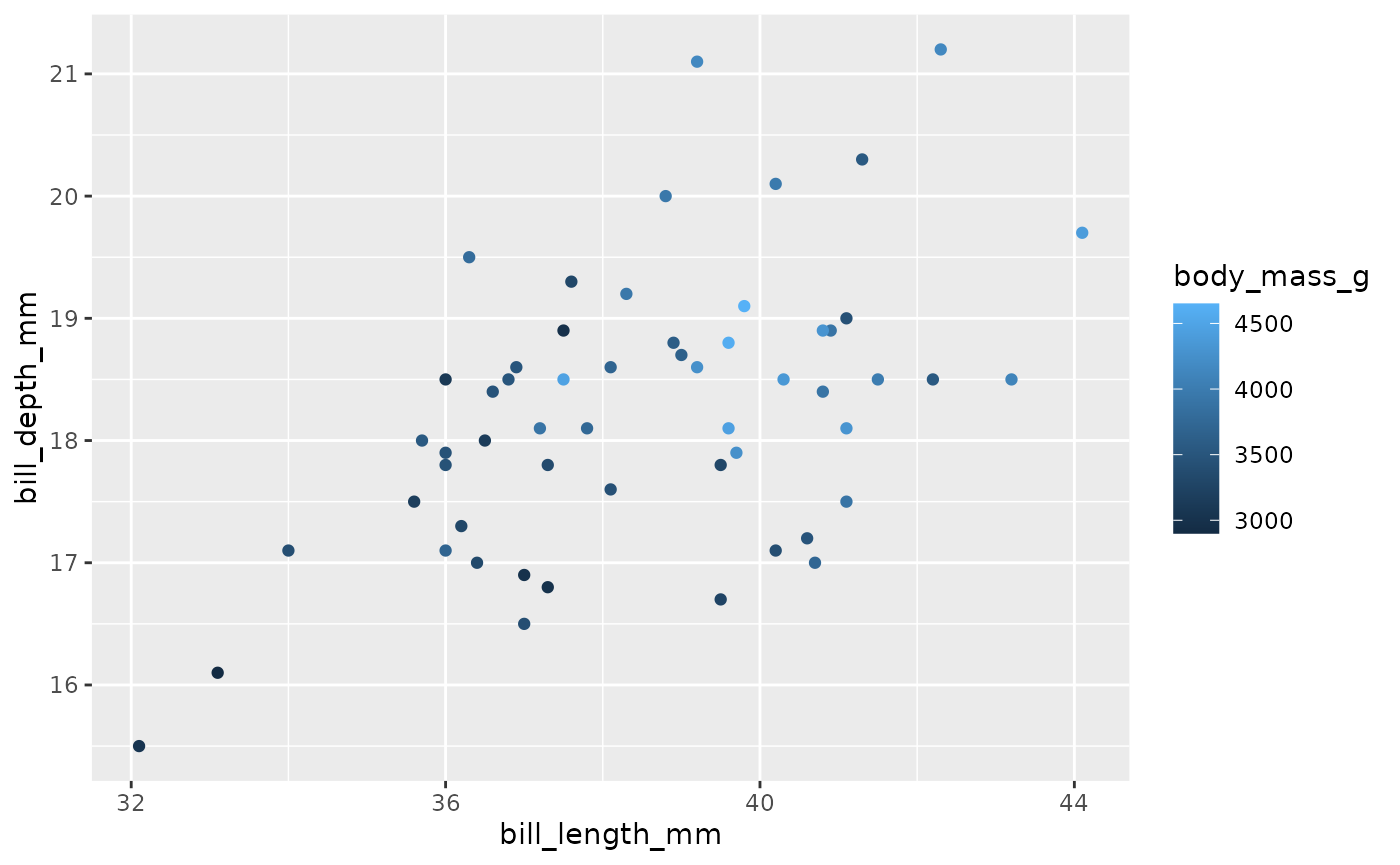
Finally, we can wrap this into a function.
plotting <- function(data){
ggplot(data, aes(x = bill_length_mm,
y = bill_depth_mm,
color = body_mass_g)) +
geom_point()
}Notice the code above is exactly as we had before, we’ve just changed our sample data set to data as an argument within a function. Normally, writing functions for plotting (and many other functions in the tidyverse) is difficult because of non-standard evaluation, but in this case we don’t have to worry about any of that.
Now, we can broadcast() the function to all of the data!
broadcasted_df = nest_df %>%
broadcast(plotting)
broadcasted_df
#> # A tibble: 5 × 4
#> # Groups: island, species [5]
#> species island data output
#> <fct> <fct> <list> <list>
#> 1 Adelie Torgersen <tibble [52 × 6]> <gg>
#> 2 Adelie Biscoe <tibble [44 × 6]> <gg>
#> 3 Adelie Dream <tibble [56 × 6]> <gg>
#> 4 Gentoo Biscoe <tibble [124 × 6]> <gg>
#> 5 Chinstrap Dream <tibble [68 × 6]> <gg>Notice we now have a new column, called output, which has all the plots! To look at the first plot, we can print it with
broadcasted_df$output[[1]]
#> Warning: Removed 1 rows containing missing values (geom_point).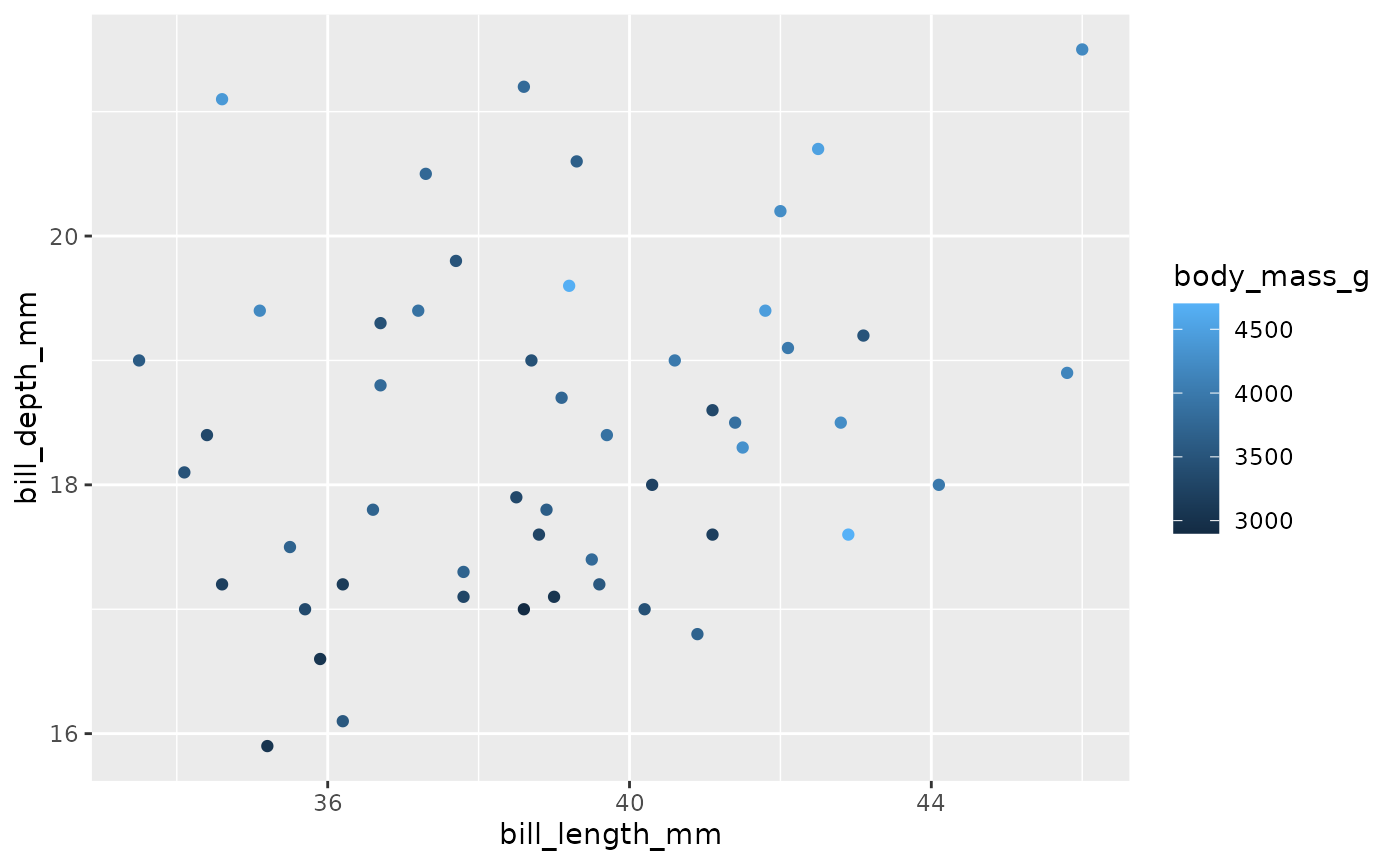
Or, similarly:
broadcasted_df %>%
pull(output) %>%
purrr::pluck(1)
#> Warning: Removed 1 rows containing missing values (geom_point).
You can also use it for modeling or other functions. For example:
tmp_df = nest_df %>%
sample_data(index = 3)
lm(body_mass_g ~ bill_length_mm * bill_depth_mm, data = tmp_df)
#>
#> Call:
#> lm(formula = body_mass_g ~ bill_length_mm * bill_depth_mm, data = tmp_df)
#>
#> Coefficients:
#> (Intercept) bill_length_mm
#> -2079.0346 92.6987
#> bill_depth_mm bill_length_mm:bill_depth_mm
#> 145.9699 -0.6616
modeling <- function(data){
lm(body_mass_g ~ bill_length_mm + bill_depth_mm, data = data)
}
nest_df %>%
broadcast(modeling) %>%
.$output
#> [[1]]
#>
#> Call:
#> lm(formula = body_mass_g ~ bill_length_mm + bill_depth_mm, data = data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm bill_depth_mm
#> -1161.63 47.79 163.14
#>
#>
#> [[2]]
#>
#> Call:
#> lm(formula = body_mass_g ~ bill_length_mm + bill_depth_mm, data = data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm bill_depth_mm
#> -3015.76 86.27 183.06
#>
#>
#> [[3]]
#>
#> Call:
#> lm(formula = body_mass_g ~ bill_length_mm + bill_depth_mm, data = data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm bill_depth_mm
#> -1622.93 80.87 120.41
#>
#>
#> [[4]]
#>
#> Call:
#> lm(formula = body_mass_g ~ bill_length_mm + bill_depth_mm, data = data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm bill_depth_mm
#> -1452.12 57.64 252.96
#>
#>
#> [[5]]
#>
#> Call:
#> lm(formula = body_mass_g ~ bill_length_mm + bill_depth_mm, data = data)
#>
#> Coefficients:
#> (Intercept) bill_length_mm bill_depth_mm
#> -356.11 23.82 158.84Instead of a list, you can also make the output a number (double).
modeling <- function(data){
model = lm(body_mass_g ~ bill_length_mm + bill_depth_mm, data = data)
model$coefficients[2] # the slope for bill_length_mm
}
nest_df %>% broadcast(modeling) %>% .$output
#> [[1]]
#> bill_length_mm
#> 47.78764
#>
#> [[2]]
#> bill_length_mm
#> 86.27406
#>
#> [[3]]
#> bill_length_mm
#> 80.86942
#>
#> [[4]]
#> bill_length_mm
#> 57.64173
#>
#> [[5]]
#> bill_length_mm
#> 23.82257
broadcast_group()
broadcast_group() allows you to also use grouping variables in the function. It is still under development so currently it only works in a more limited way. For example, you can include grouping variables as the title for plots.
df = nest_df %>%
sample_row(index = 3, type = 'list')
plotting = function(data, species, island){
ggplot(data, aes(x = bill_length_mm,
y = bill_depth_mm,
color = body_mass_g)) +
geom_point() +
labs(title = paste(species, island))
}
# example with a single plot
plotting(df$data, df$species, df$island)
nest_df = nest_df %>%
broadcast_group(plotting)
nest_df$output[[1]]
#> Warning: Removed 1 rows containing missing values (geom_point).
nest_df$output[[4]]
#> Warning: Removed 1 rows containing missing values (geom_point).